Publication | April 27, 2023
Reconstructing the landscape of gut microbial species across 29,000 diverse individuals
Get the Resource
Read the PaperThere is increasing evidence of the human gut microbiome’s link to health and disease. Exploring the composition and function of these microbial communities can help us better understand the effects of microbiota perturbation on disease development and disease treatment.
Due to the high sequencing and computational expense of whole-genome shotgun sequencing (WGS), most microbiome research has employed 16S amplicon sequencing, which has been highly successful for broad taxonomic classification but is limited in its ability to distinguish microbes at the species level. Species-level identification is essential for understanding biological mechanisms underlying the link between the microbiome and human phenotypes, and for evaluating the therapeutic potential of specific gut microbes.
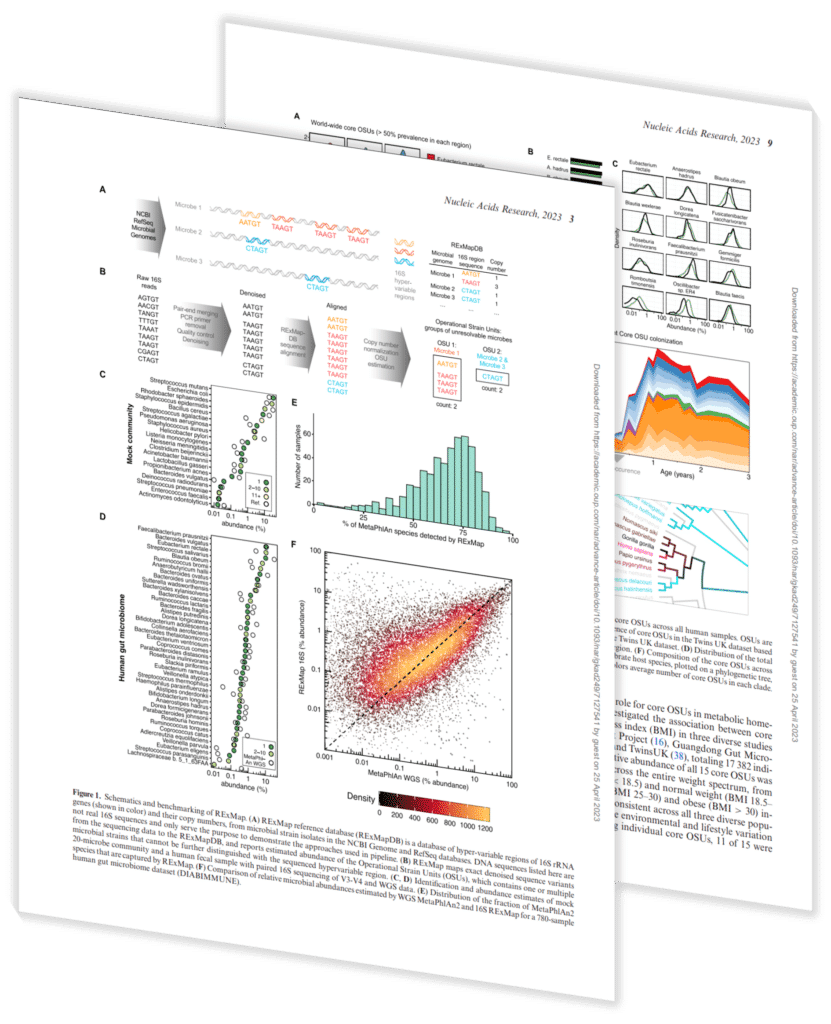
This paper, contributed to by Sapient’s scientists and published in Nucleic Acids Research, details the development of Reference-based Exact Mapping (RExMap) of microbial amplicon variants and its application in analyzing the gut microbiome of more than 29,000 diverse individuals.
RExMap analysis of 16S data captures ∼75% of microbial species identified by WGS, despite hundreds-fold less sequencing depth. The method was applied to re-analyze existing 16S data from 29,349 individuals across diverse regions around the world and reveals a detailed landscape of gut microbial species across populations and geography.
To access the full paper, click here.